General information
- Name of the project
- Friendzymes - Open Colony Picker
- Track (New Project Track / Established Project Track)
- Established Project Track

- Established Project Track
- Contact name
- Friendzymes - Jeremy Cahill; Scott Pownall
- Contact email
Proposal
Item 1
Many organizations (e.g. academic labs, startups, community labs) around the world desire low cost automation to increase the throughput of their capabilities. The success of the low cost Opentrons pipetting robot shows this. There are many other commercial laboratory automated tools that are beyond the means of many.
Colony picking robots are automated machines that can detect microbial (e.g. bacterial or yeast) colonies on agar plates, then pick and inoculate the discrete colony into liquid media for growth or replicate colonies on another agar plate. Some robots can distinguish between fluorescent and non-fluorescent colonies. Automation of the process of colony picking allows high throughput and frees up expensive manual labor.
Item 2
Friendzymes received round 1 funding in the Established Project Track for our project whose goal is to create a low cost open automated colony picking robot to facilitate molecular cloning of assembled DNA parts from Freegenes, iGEM, and other open wetware sources as well as for colony screening of strains for desired assemblies and behavior.
The focus of our round 1 application was on developing some of the subsystems required to realize a complete automated colony picking robot. This included designing and building the Colony Picking Head Subsystem, the Raspberry Pi-based FluoPi Imaging Subsystem and ideation of the completed robot.
Phase 2 goals are to integrate those Phase 1 sub-systems along with some new subsystems to be designed and built in Phase 2. All of these subsystems will be integrated with the OpenWorkstation cartesian system using the Klipper 3D framework. All of the subsystems for the final colony picking robot are:
- Cartesian System based on OpenWorkstation
- Adapted FloPi subsystem
- Bright field and fluorescent plate imaging
- Discrete colony identification
- Map agar surface height
- Colony Pick Head Subsystem
- Sterilization Unit
- Filament Cutting Subsystem
- Head Transport
- Agar Plate Management Subsystem
- Agar Plate Stack Holding Rack
- Agar Plate Transport Mechanism
- Lid Lifter Mechanism
- SBS Deep Well Plate Management
- Software
Item 3
Our open source automated colony picker is to be used by microbiology and synthetic biology labs that routinely run protocols where imaging, plating, replicating and culturing bugs take up a significant amount of time due to the scale or the yield of the prescribed protocol. Typically, there is an inflection point for some labs where hiring more warm bodies exceeds the cost of automating the process. However, due to the high price of commercial pickers, the cost of acquisition can often exceed the capital expenditure requirements of some labs despite the operational scale advantages of having such machines, not to mention the cost and difficulty in setting up and maintaining such machines. In conversations with some partner labs around the world, this can happen where personnel are hired on a per-project contract basis. Thus, a niche exists wherein the most following conditions apply:
- There are minimal yet highly skilled personnel operating the lab.
- The laboratory is located in a remote region of significance where a vast majority of samples have to be processed onsite, e.g. field labs.
- The costs of acquisition of a commercial colony picking unit vastly exceeds the capital expenditure ability of the laboratory.
- The laboratory is located in a remote site where setup and maintenance is difficult.
- The required yield is vastly less than a commercial picking unit yet more than what the lab personnel can do manually.
By using this unit, time spent picking is converted from manpower time to machine time therefore increasing the productivity of the said lab at minimal depreciated cost to the lab itself.
Due to the conditions above, there are a few requirements that need to be satisfied. First, the colony picker should be field maintainable or at least easily transportable without the use of heavy lifting devices. Second, manual user intervention in the picking process must be minimized to realize the maximum advantage of having the machine. Third, all parts of the machine should be easily procurable in most markets.
Item 4
The focus of our round 1 application was on developing some of the subsystems required to realize a complete automated colony picking robot and run through a number of rounds of ideation of the completed robot to identify what additional subsystems are required and how they would integrate with the OpenWorkstation cartesian system.
Majority of the software work done was testing, integrating, and validating OpenCFU with Python bindings for integration into PAML as a separate specialization. As part of the work completed, a boilerplate specialization for the OT-2 as a base was made for later generalization into a generic cartesian gantry system. This would later allow for PAML protocols to natively use the colony picker as a protocol execution component. PAML thus can act as the coordinator of all the movements of the robot. Integration and multi-execution tests are ongoing.
For firmware development, we elected to go with Klipper for the motion control and hardware control of the robot. This is due to the extensibility of Klipper with many 3d printer boards as well as the ability to update the firmware on the fly via the printer.cfg facility. Macros could also be developed for the “plate elevator” to align and calibrate the system for different plate sizes and form factors. For now, work is being done to reverse engineer and reduce the pertinent thermal safeties of the firmware in a safe manner that should not impede robot operation.
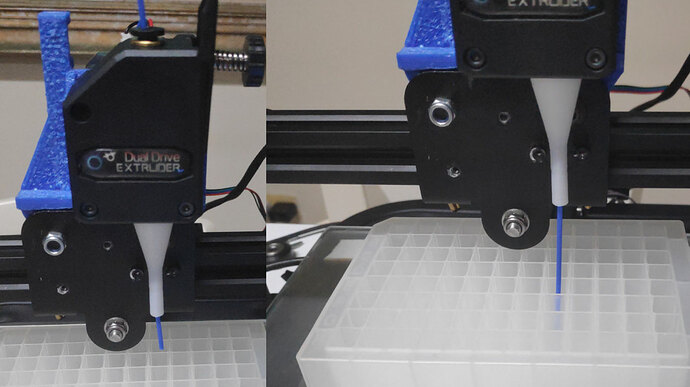
Hardware subsystems in round 1 included designing and building the Colony Picking Head Subsystem, the Raspberry Pi-based FluoPi Imaging Subsystem and using OpenCV to identify discrete colonies as well as map the agar surface.
Although not originally included in the round 1 application, we designed, implemented and tested a 3D printed vacuum-based lid lifting mechanism for lifting lids off of the agar plates.
Item 5
Round 1 Contributors continue to participate in the project:
- The Friendzymes Contributors
- Jeremy Cahill, USA
- Isaac Larkin, USA
- Sarah Ware, USA
- Isaac Nuñez – FluoPi, Chile
- Scott Pownall, Canada
- Homer Sajonia II, Philippines
- Jacob Segarra, USA
- Friendzymes Advisor
- Sebastian Eggert – OpenWorkstation, Germany
New Contributors include the following cohorts:
- The Friendzymes Contributors
- Ian Caven, Canada (OSN)
- Open Insulin
- Moses Apostol, USA
- Joshua Harrell, USA
- Yann Huon (PI), France
- OpenTrons Engineering
- Nick Diehl, USA
As members of the Open Insulin project, Moses Apostol, Joshua Harrell, and Yann Huon (PI) share substantial interests with Friendzymes in the development of open colony picking methods. This includes the application of computer vision to imaging tasks with OpenTrons hardware and OpenCFU software. At the nexus of their work with Open Insulin and this colony picker project, Moses and Joshua have laid the groundwork for OT-2 calibration and colony picking pipeline automation, taking as input a colony plate image with the end goal of executing transfer protocols.
Nick Diehl is an Applications Engineer at OpenTrons based in Brooklyn, New York. Nick graduated from Vanderbilt University with a Biomedical Engineering degree in 2018. He joined Opentrons in 2019 to deliver bespoke solutions to OT-2 users around the world.
Ian Caven is a software engineer and active member of Open Science Network’s community lab. He has extensive experience with 3D printing and developing CAD files using open source tools like OpenSCAD. Ian and Scott collaborated to design and print a working lid lifting device. Ian realized the design using OpenSCAD.
Item 6
Friendzymes’ mission is to democratize and globally distribute the means of biotechnological production. The goal of this project remains, as in Round 1, to design and build a low cost cartesian-based open automated colony picking robot using the Raspberry Pi and associated camera system.
Over the course of research, design, and development during Round 1, we successfully created the Colony Picking Head and sterilizing unit and adapted the FluoPi imaging device to integrate with OpenCV for detecting the location of discrete colonies on agar plates.
In this latter phase of our prospective GOSH Collaborative Development Program Phase 2 funding, we aim to extend the work of Round 1 by creating additional subsystems as defined elsewhere in this application and integrating all of the subsystems in a cartesian framework based on OpenWorkstation. We expect we will need a number of iterations in the design, build, test, learn cycle to ensure all components work as we envision.
Item 7
Please find our Gantt chart attached as PNG and PDF below.
Friendzymes_GOSH_Collaborative_Development_Phase_02_Gantt.pdf
Item 8
Please find our Budget (BOM) attached as a spreadsheet below.
Total Budget Request: 17874.78 USD
Friendzymes_GOSH_Collaborative_Development_Phase_02_Budget.xlsx
Item 9
As in Round 1:
- Friendzymes’ main tool for communication remains our Discord channel.
- Friendzymes maintains a public GitHub organization, where all project materials will be disseminated.
- Friendzymes continues as a multi-focal project, with individual team members continuing to organize regularly scheduled team calls and thematic development sessions.
- Updated links can always be found at https://linktr.ee/friendzymes.
Closing Remarks
Friendzymes would like to thank the Round 1 reviewers for giving this project the opportunity to be realized and the Round 2 reviewers for their consideration of this application.